Cancer Biomarker Discovery (CaBiD)
A GUI tool for investigating DEGs and biomarker discovery


I am currently working on developing a GUI for performing Differential Gene Expression Analysis and Gene Set Enrichment Analysis on Microarray data generated from cancer patients. The tool will allow researchers to perform the analysis on their own data and identify potential biomarkers for cancer diagnosis and treatment. The GUI was developed in Python using PyQt6 and I am currently working on integrating a backend in R to perform the analysis with the limma and DESeq2 packages. I am also working on a web application for the tool using Flask.
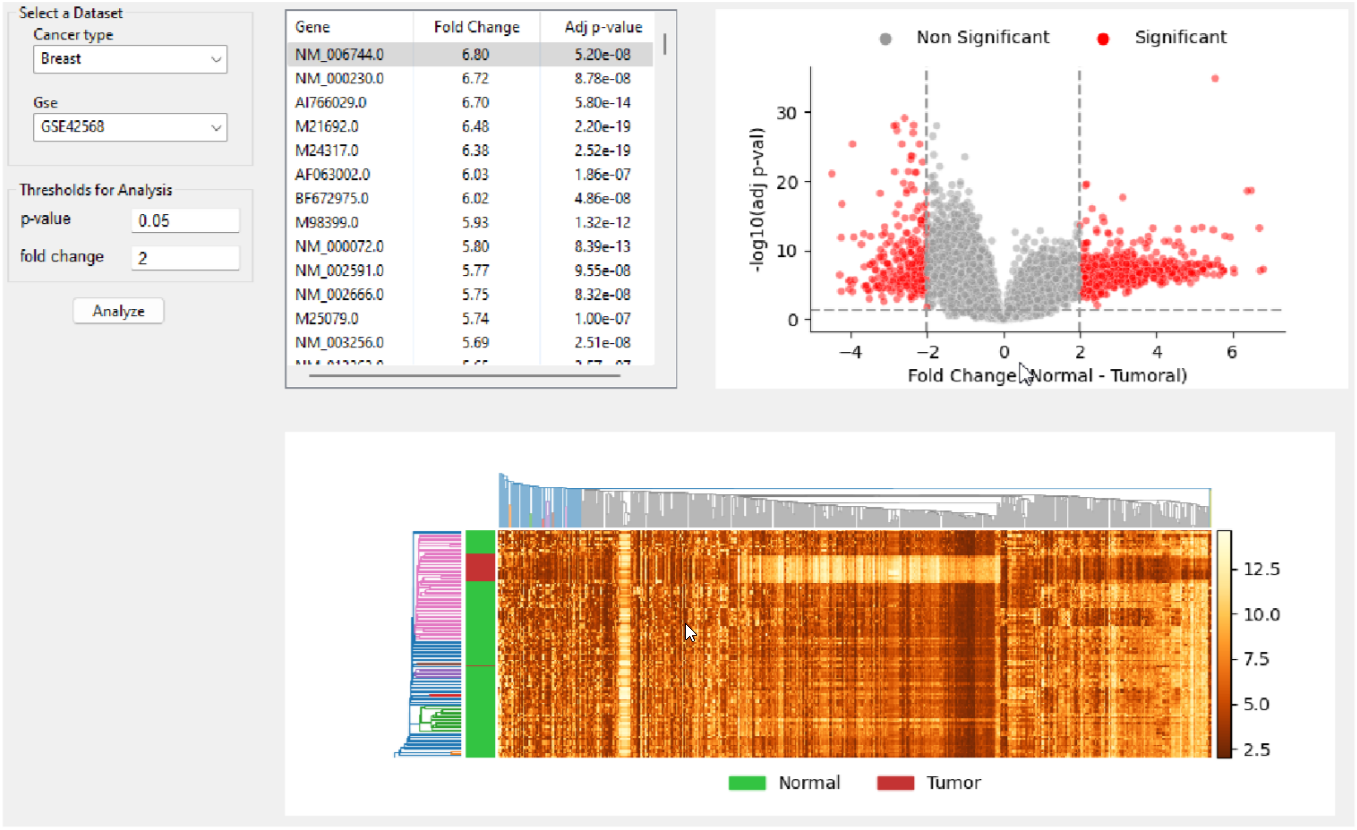
The goal of this project is to develop a web application to investigate variations in gene expression across various cancer types. Datasets selected from GEO (Gene Expression Omnibus) and CuMiDa (Curated Microarray Database) will be preprocessed and curated in a SQLite database. The wxPython package will then be used to develop a GUI application that will generate visualizations of the user-selected dataset. The software will identify key differences in gene expression between healthy controls and tumoral samples across various cancer types. The GUI will include heatmaps and volcano plots to visualize the differences in gene expression, along with a table of the significantly differentially expressed genes. To find out more about the project, please take a look at the project report. You can also take a look at the project design document.